Prior-aware Neural Network for Partially-Supervised Multi-Organ Segmentation
[ICCV’19]
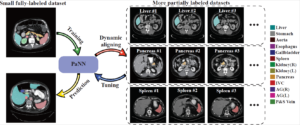 Accurate multi-organ abdominal CT segmentation is essential to many clinical applications such as computer-aided intervention. As data annotation requires massive human labor from experienced radiologists, it is common that training data are partially labeled, e.g., pancreas datasets only have the pancreas labeled while leaving the rest marked as background. However, these background labels can be misleading in multi-organ segmentation since the “background” usually contains some other organs of interest. To address the background ambiguity in these partially-labeled datasets, we propose Prior-aware Neural Network (PaNN) via explicitly incorporating anatomical priors on abdominal organ sizes, guiding the training process with domain-specific knowledge. More specifically, PaNN assumes that the average organ size distributions in the abdomen should approximate their empirical distributions, prior statistics obtained from the fully-labeled dataset. As our training objective is difficult to be directly optimized using stochastic gradient descent, we propose to reformulate it in a min-max form and optimize it via the stochastic primal-dual gradient algorithm. PaNN achieves state-of-the-art performance on the MICCAI2015 challenge “Multi-Atlas Labeling Beyond the Cranial Vault”, a competition on organ segmentation in the abdomen. We report an average Dice score of 84.97%, surpassing the prior art by a large margin of 3.27%.
Accurate multi-organ abdominal CT segmentation is essential to many clinical applications such as computer-aided intervention. As data annotation requires massive human labor from experienced radiologists, it is common that training data are partially labeled, e.g., pancreas datasets only have the pancreas labeled while leaving the rest marked as background. However, these background labels can be misleading in multi-organ segmentation since the “background” usually contains some other organs of interest. To address the background ambiguity in these partially-labeled datasets, we propose Prior-aware Neural Network (PaNN) via explicitly incorporating anatomical priors on abdominal organ sizes, guiding the training process with domain-specific knowledge. More specifically, PaNN assumes that the average organ size distributions in the abdomen should approximate their empirical distributions, prior statistics obtained from the fully-labeled dataset. As our training objective is difficult to be directly optimized using stochastic gradient descent, we propose to reformulate it in a min-max form and optimize it via the stochastic primal-dual gradient algorithm. PaNN achieves state-of-the-art performance on the MICCAI2015 challenge “Multi-Atlas Labeling Beyond the Cranial Vault”, a competition on organ segmentation in the abdomen. We report an average Dice score of 84.97%, surpassing the prior art by a large margin of 3.27%.
Thoracic Disease Identification and Localization with Limited Supervision
[CVPR’18]
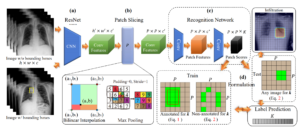 Accurate identification and localization of abnormalities from radiology images play an integral part in clinical diagnosis and treatment planning. Building a highly accurate prediction model for these tasks usually requires a large number of images manually annotated with labels and finding sites of abnormalities. In reality, however, such annotated data are expensive to acquire, especially the ones with location annotations. We need methods that can work well with only a small amount of location annotations. To address this challenge, we present a <strong>unified approach</strong> that simultaneously performs <strong>disease identification and localization</strong> through the same underlying model for all images. We demonstrate that our approach can effectively leverage both class information as well as limited location annotation, and significantly outperforms the comparative reference baseline in both classification and localization tasks.
Accurate identification and localization of abnormalities from radiology images play an integral part in clinical diagnosis and treatment planning. Building a highly accurate prediction model for these tasks usually requires a large number of images manually annotated with labels and finding sites of abnormalities. In reality, however, such annotated data are expensive to acquire, especially the ones with location annotations. We need methods that can work well with only a small amount of location annotations. To address this challenge, we present a <strong>unified approach</strong> that simultaneously performs <strong>disease identification and localization</strong> through the same underlying model for all images. We demonstrate that our approach can effectively leverage both class information as well as limited location annotation, and significantly outperforms the comparative reference baseline in both classification and localization tasks.
Block-Circulant Matrix-Based Deep Learning Systems
[AAAI’18][ICML’17][MICRO’17][ICCAD’17][FPGA’18]
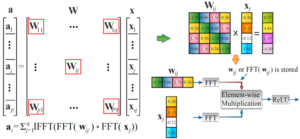
The rapidly expanding model size in deep learning systems is posing a significant restriction on both the computation and weight storage, for both inference and training, and on both high-performance computing systems and low-power embedded system and IoT applications. In order to overcome these limitations, we propose a holistic framework of incorporating structured matrices (block-circulant matrices) into deep learning systems, and could achieve (i) simultaneous reduction on weight storage and computational complexities, (ii) simultaneous speedup of training and inference, and (iii) generality and fundamentality that can be adopted to both software and hardware implementations, different platforms, and different neural network types, sizes, and scalability.
Besides algorithm-level achievements, our framework has (i) a solid theoretical foundation to prove that our approach will converge to the same “effectiveness” as deep learning without compression; (ii) platform-specific implementations and optimizations on smartphones, FPGAs, and ASIC circuits. We demonstrate that our smartphone-based implementation achieves the similar speed of GPU and existing ASIC implementations on the same application. Our FPGA-based implementations for deep learning systems and LSTM networks could achieve 10X+ energy efficiency improvement compared with state-of-the-art, and even higher energy efficiency gain compared with IBM TrueNorth. Our proposed framework can achieve 3.5 TOPS performance in FPGAs and is the first to enable nano-second level recognition speed for image recognition tasks.
SC-DCNN: Deep Convolutional Neural Networks using Stochastic Computing
[ICRC’16][ICCD’16][ASPDAC’17][ASPLOS’17][DATE’17][IJCNN’17]
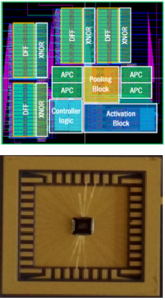

Stochastic Computing (SC), which uses a bit-stream to represent a number within [-1, 1] by counting the number of ones in the bit-stream, has the high potential for implementing DCNNs with high scalability and ultra-low hardware footprint. Since multiplications and additions can be calculated using AND gates and multiplexers in SC, significant reductions in power (energy) and hardware footprint can be achieved.
In this project, we propose SC-DCNN, the first comprehensive design, and optimization framework of SC-based DCNNs, using a bottom-up approach. We first design the function blocks that perform the basic operations in DCNN, including inner product, pooling, and activation function. Then we propose four designs of feature extraction blocks, which are in charge of extracting features from input feature maps, by connecting different basic function blocks with joint optimization. Putting all together, SC-DCNN is holistically optimized to minimize area and power (energy) consumption while maintaining high network accuracy.